|
I am a PhD Candidate at the Electrical Engineering department of the Technion, jointly supervised by Prof. Tomer Michaeli and Prof. Yoav Shechtman. Prior to that, I did my bachelors in Biomedical Engineering also at the Technion. My research lie at the intersection of computational imaging, computer vision, and machine learning, and their application to the domain of bioimaging/microscopy. During my PhD, I mostly worked on next generation cameras employing Computational Optics and deep neural networks to recover physical properties of the imaged scene. Specifically, I worked on optimizing reconstruction from challenging measurements, on end-to-end learning of optics and image processing for precise depth estimation, and more recently on visualizing and quantifying reconstruction uncertainty in ill-posed inverse problems in imaging. Throughout my studies, I had the privlige of frequently collaborating with Dr. Daniel Freedman from Verily research center in Haifa. |

|
|
Sep 26, 2024 - Posterior Trees was accepted to NeurIPS 2024. Jun 16, 2024 - I gave an invited talk on visualizing uncertainty at the HUJI Vision Seminar. Jun 14, 2024 - I gave a virtual invited talk on visualizing uncertainty at the AstraZeneca Center for Artificial Intelligence. May 27, 2024 - I gave an invited talk on visualizing uncertainty at the AIM24 conference. May 03, 2024 - Our High-throughput work in collaboration with Sartorius was accepted to Nature Communications. Mar 13, 2024 - The large-FOV DeepSTORM3D paper now appears in Science Advances. Feb 27, 2024 - PPDE was accepted to CVPR 2024. Sep 21, 2023 - NPPC was accepted to NeurIPS 2023. Jun 01, 2023 - I gave an invited talk on Intelligent Microscopy at the AI for Scientific Data Analysis Mini-conference. Dec 21, 2022 - The optimal Multicolor 3D PSF work now appears in Intelligent Computing. Dec 15, 2022 - Our Compact PSF Engineering patent is now published. May 20, 2022 - Our cell-cycle dependent chromatin dynamics work now appears in iScience. Feb 09, 2022 - Our ICCP 2021 work won an Excellent Paper Award at the MLIS-TCE Conference. Oct 04, 2021 - I started a Research Scientist Intern position at Verily (=Google Life Sciences). Jul 19, 2021 - Deep-ROCS now appears in Optics Express. May 27, 2021 - QAFKA now appears in The Journal of Physical Chemistry B. May 24, 2021 - The Liquid-immersed Optics paper now appears in Nature Communications. Apr 15, 2021 - The collaborative project ZeroCostDL4Mic now appears in Nature Communications. Feb 28, 2021 - Our Multi-PSF Learning work was accepted to ICCP 2021 and selected for a special issue of TPAMI. Feb 17, 2021 - I received the Jacobs-Qualcomm Fellowship for Outstanding Ph.D. Students 2020-2021. Oct 19, 2020 - DeepSTORM3D was covered in a Nature Methods Technology Feature on smart microscopy. Aug 23, 2020 - I received the VATAT Prize for Research in Applied Data Science 2019. Jun 16, 2020 - DeepSTORM3D was covered in YNET, Jerusalem Post, American Technion Society, and Glocalist! Jun 14, 2020 - DeepSTORM3D was featured on the cover of the Lorry I. Lokey Interdisciplinary center Annual Report. Jan 25, 2020 - DeepSTORM is the Top-cited article in Optica from 2018 with over 200 citations! Jan 09, 2019 - DeepSTORM3D won the Best Poster Award at QBI 2019. Jun 20, 2018 - DeepSTORM won the Lev-Margulis Prize at ISM 2018. May 31, 2018 - DeepSTORM was covered in a Nature Methods Research Highlight. |
|
|
 |
Elias Nehme, Rotem Mulayoff, Tomer Michaeli Advances in Neural Information Processing Systems (NeurIPS), 2024 [paper] It is possible to learn tree-based hierarchical representations of reconstruction uncertainty in imaging inverse problems. |
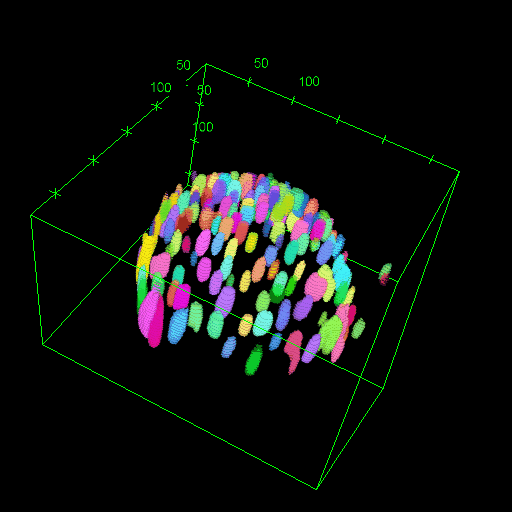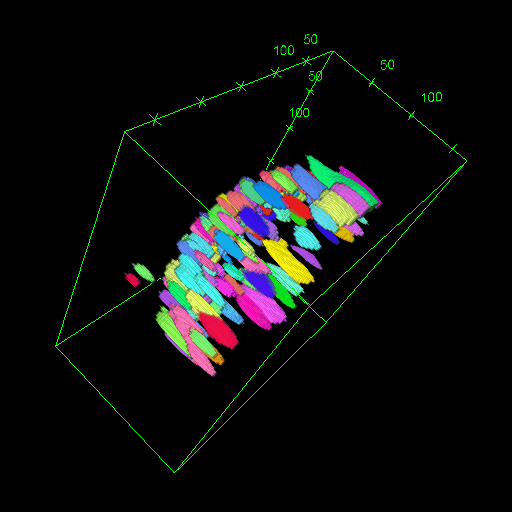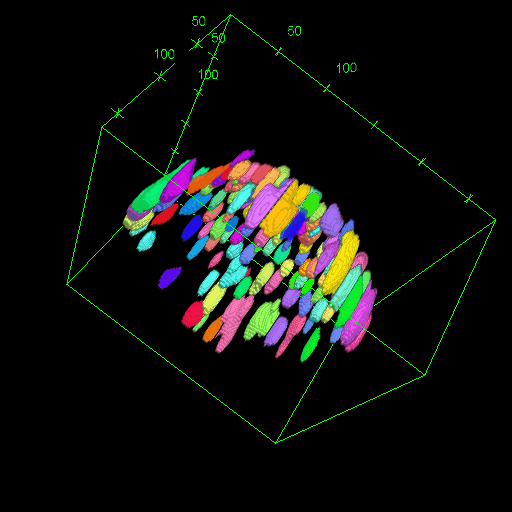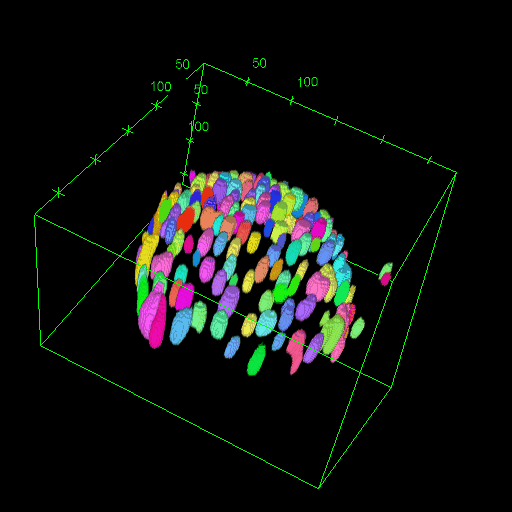 |
Nadav Opatovski*, Elias Nehme*, Noam Zoref, Ilana Barzilai, Reut Orange-Kedem, Boris Ferdman, Paul Keselman, Onit Alalouf, Yoav Shechtman Nature Communications, 2024 [paper] / [code] Together with Sartorius we have shown that it is possible to make high-throughput microscopes image in 3D much faster. |
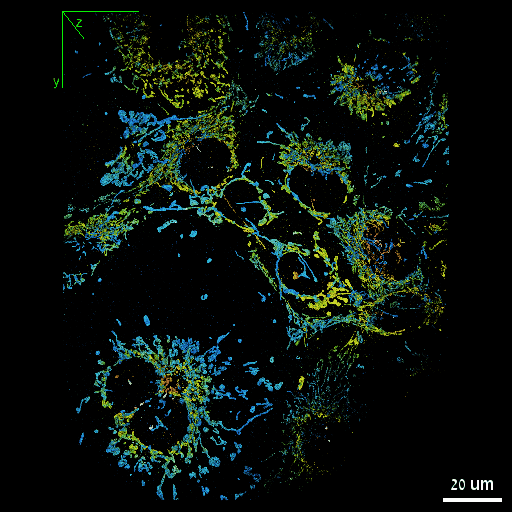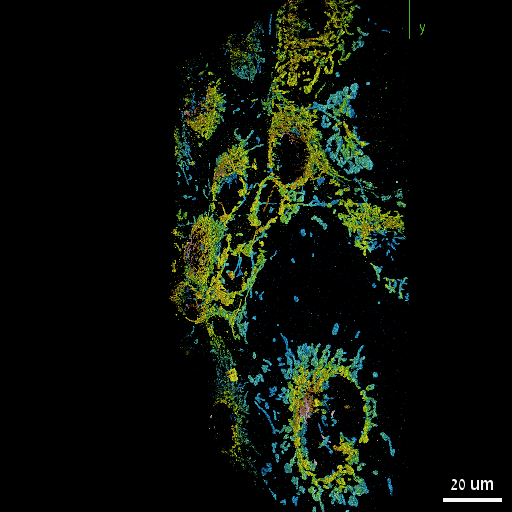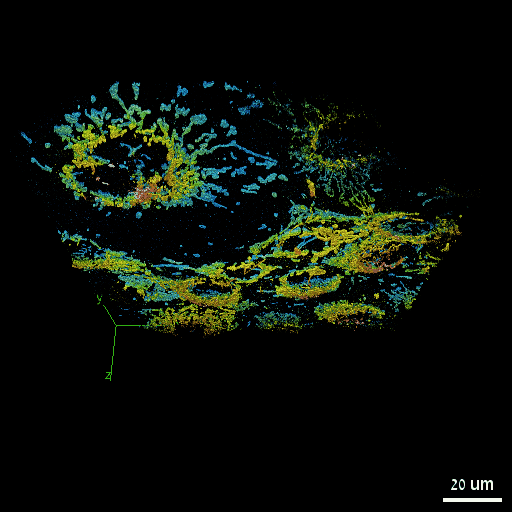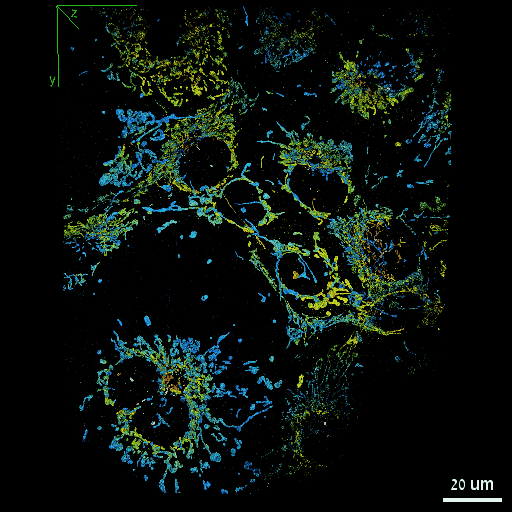 |
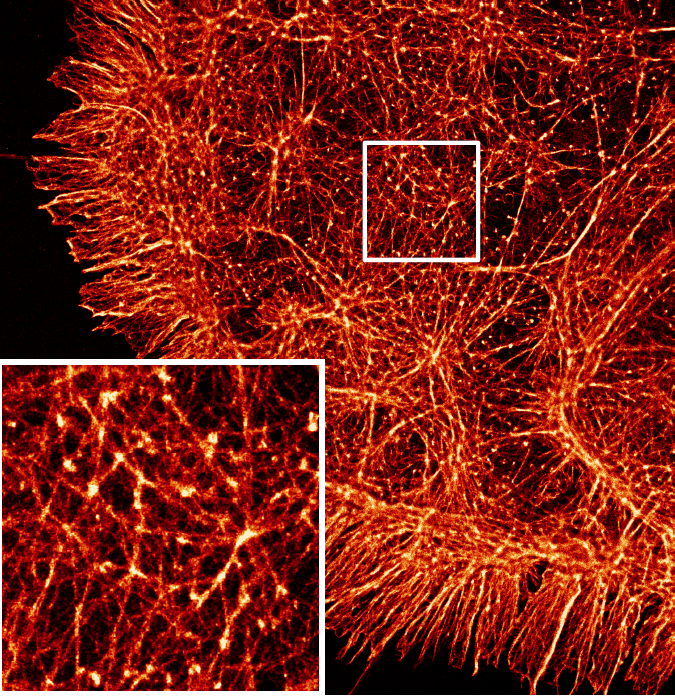
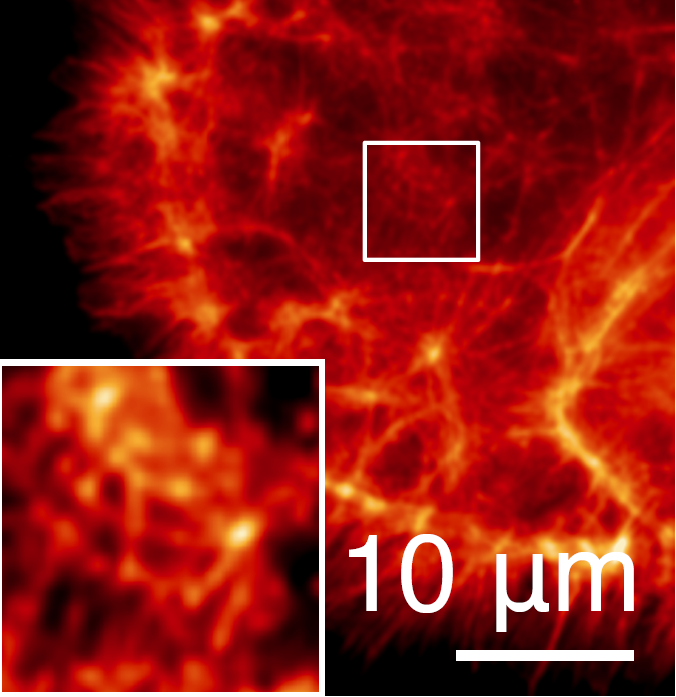
Dafei Xiao, Reut Orange-Kedem, Nadav Opatovski, Amit Parizat, Elias Nehme, Onit Alalouf, Yoav Shechtman Science Advances, 2024 [paper] / [code] It is possible to extend DeepSTORM3D to large FOVs by efficiently modelling the spatially-varying point spread function. |
|
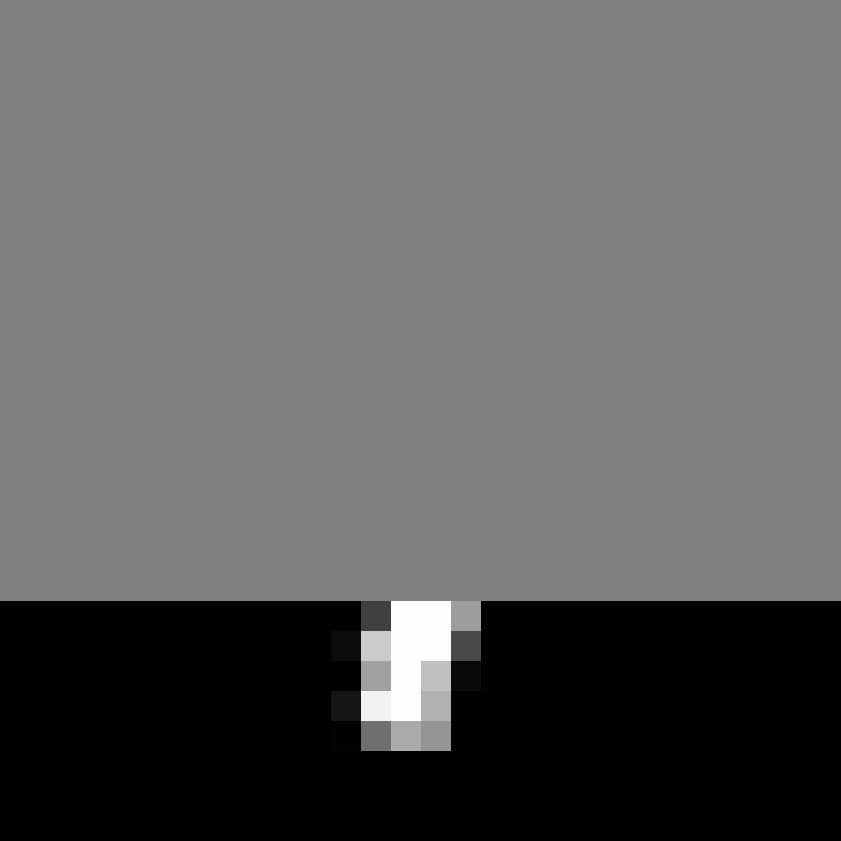
Omer Yair, Elias Nehme, Tomer Michaeli IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2024 [paper] / [code] / [project webpage] It is possible to visualize reconstruction uncertainty in imaging inverse problems by learning informative low dimensional projections of the posterior distribution. |
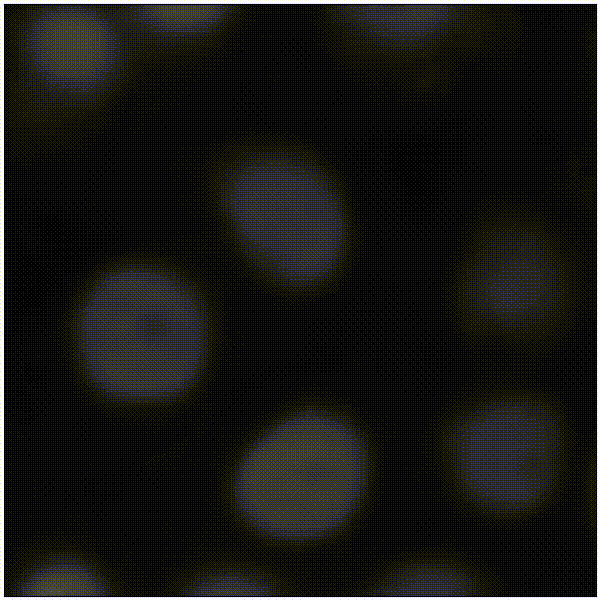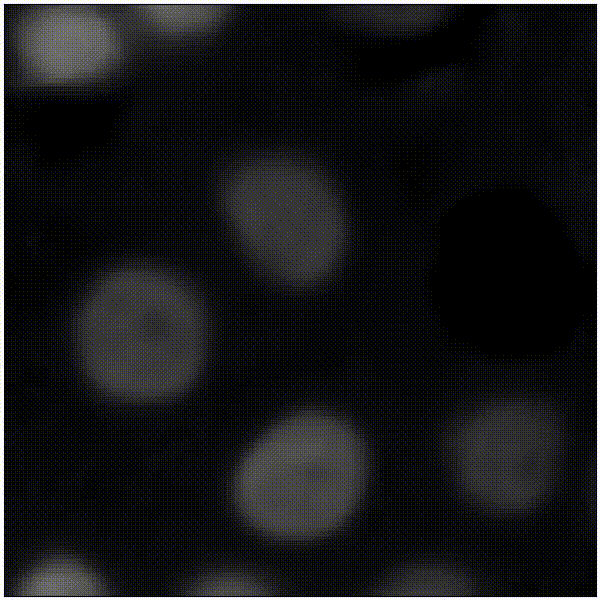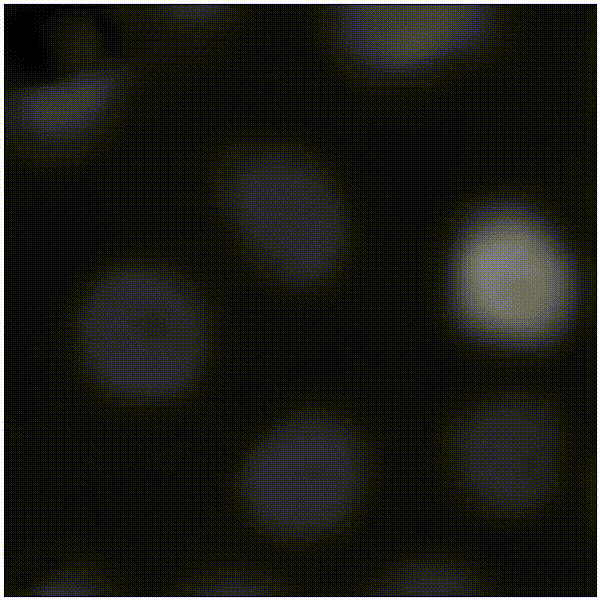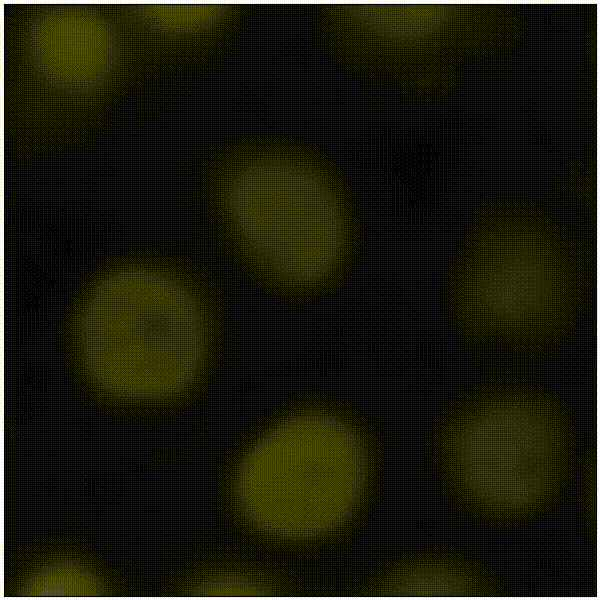 |
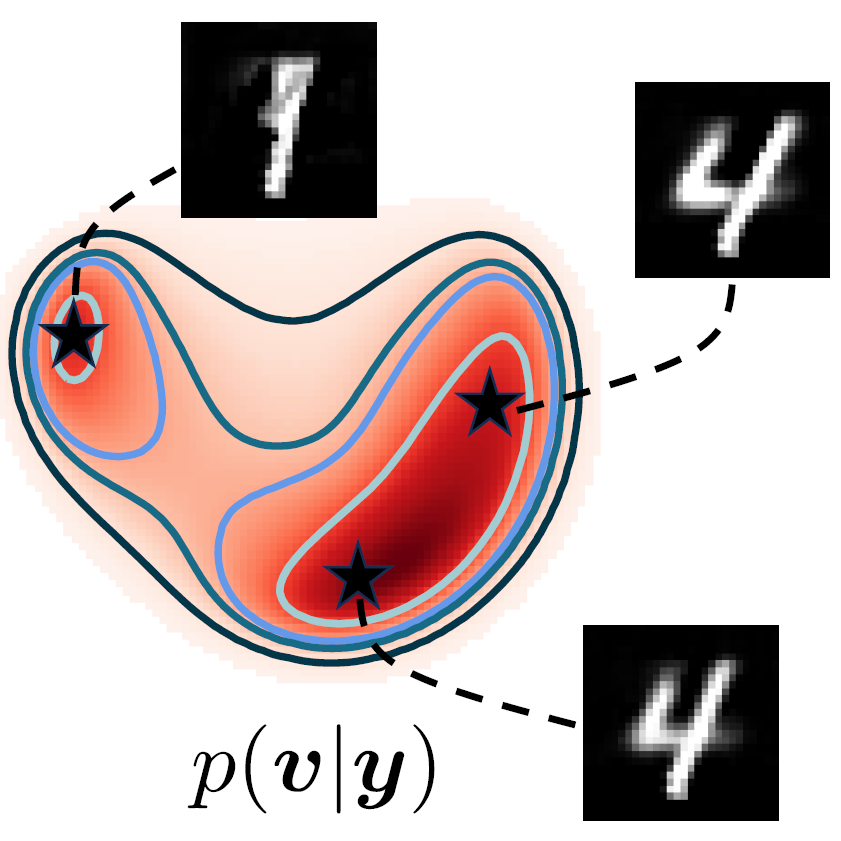
Elias Nehme, Omer Yair, Tomer Michaeli Advances in Neural Information Processing Systems (NeurIPS), 2023 [paper] / [code] / [video] / [project webpage] It is possible to learn the input-adaptive top posterior PCs and visualize reconstruction uncertainty in imaging inverse problems. |
|
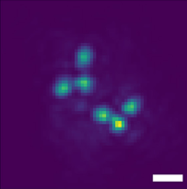
Ofri Goldenberg, Boris Ferdman, Elias Nehme, Yael Shalev Ezra, Yoav Shechtman Intelligent Computing, 2022 [paper] Multicolor 3D PSF engineering enables measuring distance between 2 fluorescently tagged DNA loci in yeast cells. |
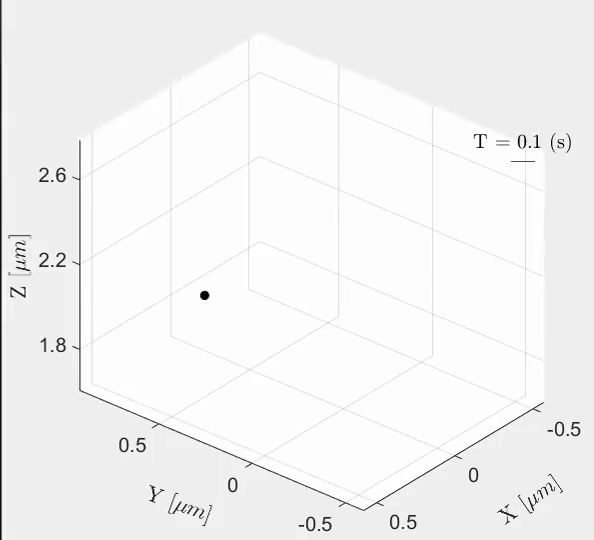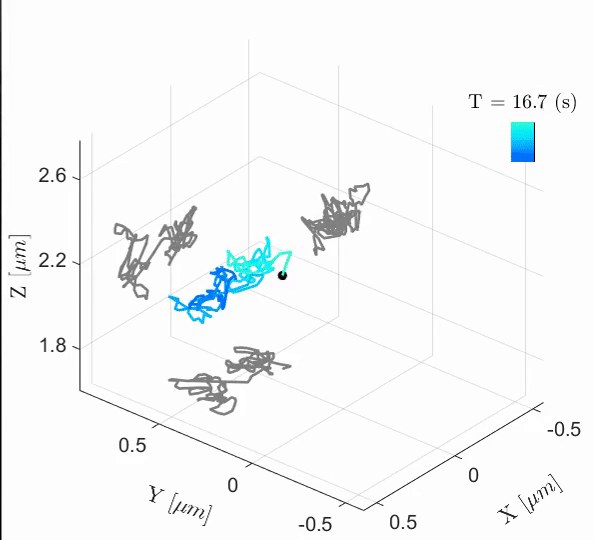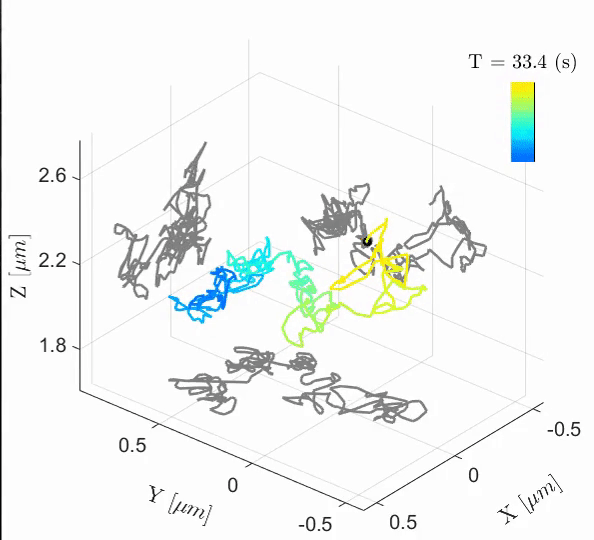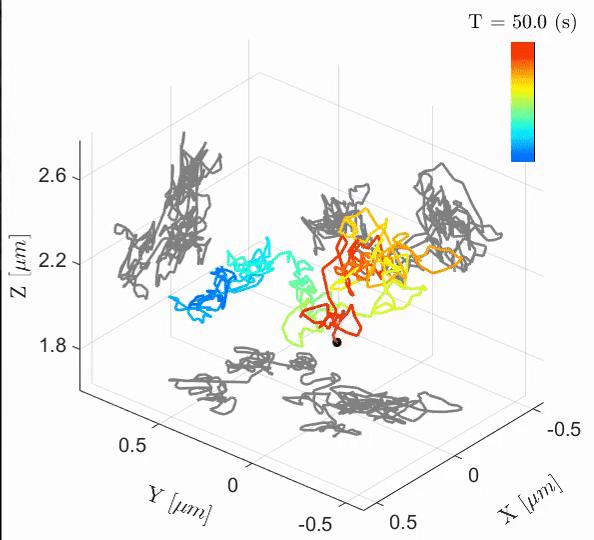 |
Tal Naor, Yevgeni Nogin, Elias Nehme, Boris Ferdman, Lucien E. Weiss, Onit Alalouf, Yoav Shechtman iScience, 2022 [paper] / [code] Cell-cycle-dependent chromatin dynamics in live cells can be quantified with high spatiotemporal resolution using end-to-end learned optics and image processing. |
|
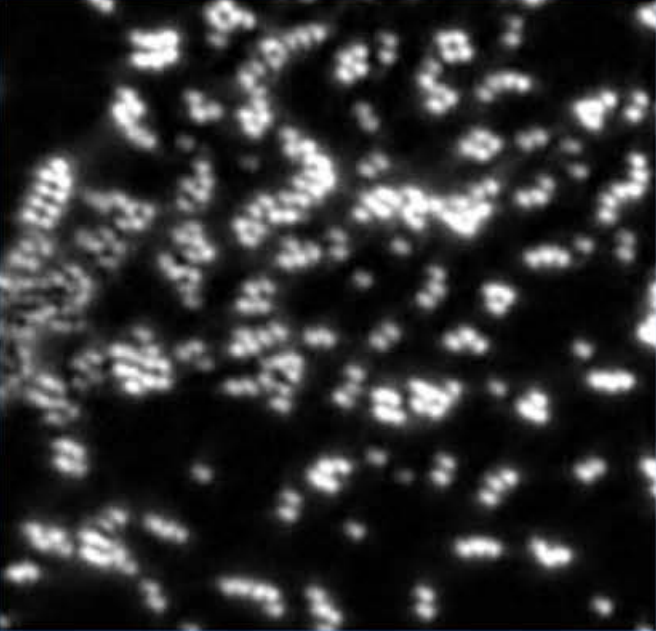
Alon Saguy, Felix Jünger, Aviv Peleg, Boris Ferdman, Elias Nehme, Alexander Rohrbach, Yoav Shechtman Optics Express, 2021 [paper] Efficient numerical combination of a set of differently illuminated images retrieves high-frequency information from a small number of speckle images. |
|
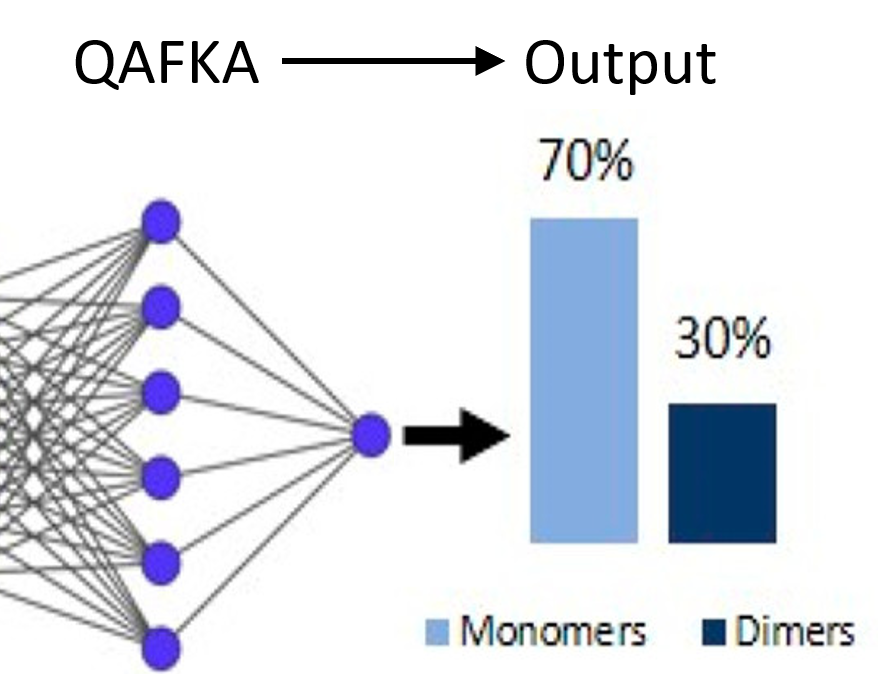
Alon Saguy, Tim N. Baldering, Lucien E. Weiss, Elias Nehme, Christos Karathanasis, Marina S. Dietz, Mike Heilemann, Yoav Shechtman The Journal of Physical Chemistry B, 2021 [paper] / [code] Extracting and analyzing blinking events of fluorophore clusters with neural networks enables quantification of oligomerization level in membrane receptors. |
 |
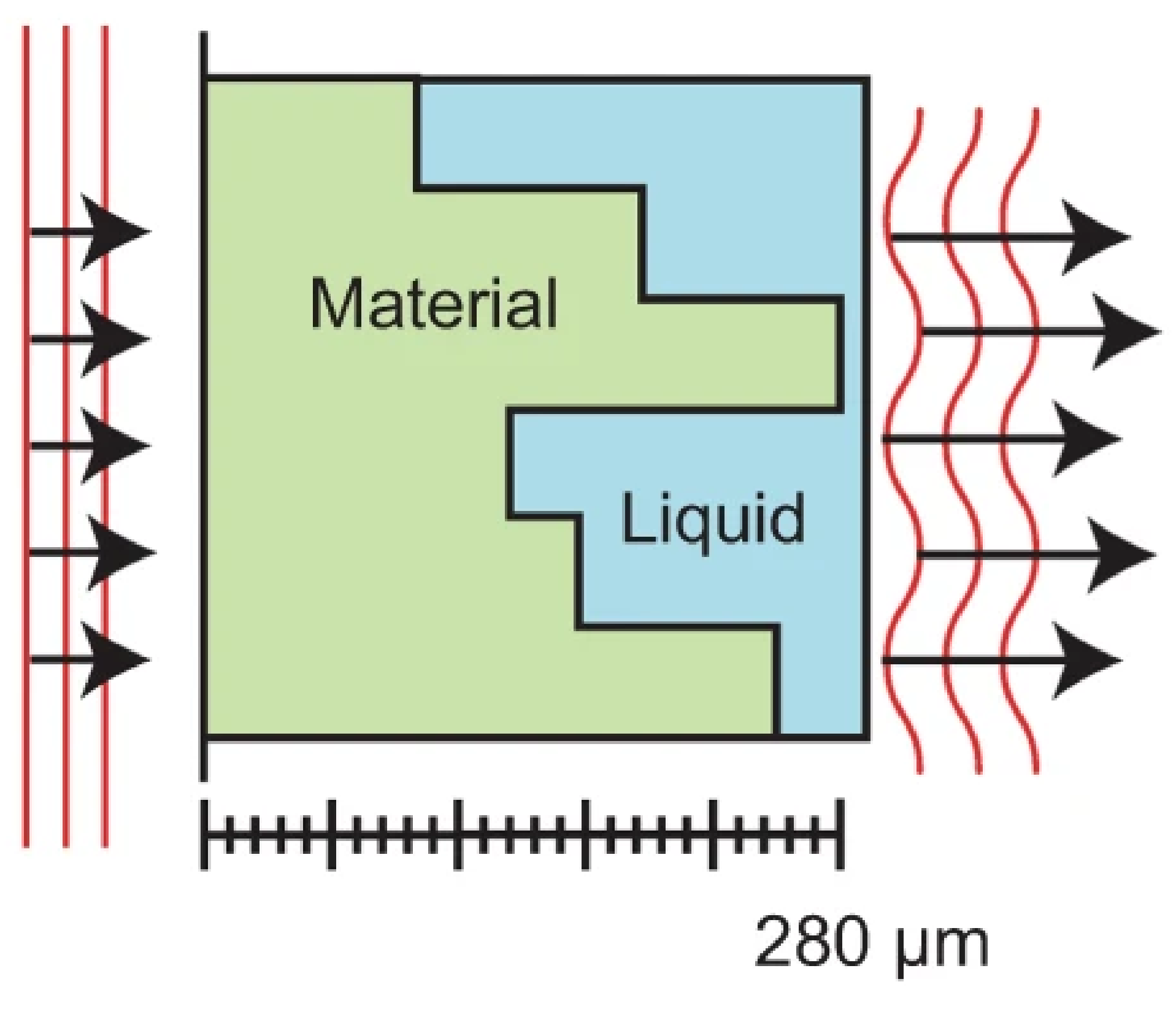
Reut Orange-Kedem, Elias Nehme, Lucien E. Weiss, Boris Ferdman, Onit Alalouf, Nadav Opatovski, Yoav Shechtman Nature Communications, 2021 [paper] Diffractive Optical Elements fabrication can be made orders of magntiude cheaper by immersing 3D printed designs in a near-index-matched solution. |
 |
Elias Nehme*, Boris Ferdman*, Lucien E. Weiss, Tal Naor, Daniel Freedman, Tomer Michaeli, Yoav Shechtman ICCP, 2021 (Selected for Special Issue of TPAMI) [paper] / [video] Given a budget of signal photons, we learn the optimal wavefront codes for recovering the depth of nanometric point sources under challenging conditions. |
|
Lucas Von Chamier, Romain F. Laine, Johanna Jukkala, Christoph Spahn, Daniel Krentzel, Elias Nehme, Martina Lerche, Sara Hernández-Pérez, Pieta K. Mattila, Eleni Karinou, Séamus Holden, Ahmet Can Solak, Alexander Krull, Tim-Oliver Buchholz, Martin L. Jones, Loic A. Royer, Christophe Leterrier, Yoav Shechtman, Florian Jug, Mike Heilemann, Guillaume Jacquemet, Ricardo Henriques Nature Communications, 2021 [paper] / [code] Self-explanatory collection of Colab notebooks to simplify access and use of deep learning in microscopy. |
 |
Racheli Gordon-Soffer, Lucien E. Weiss, Ran Eshel, Boris Ferdman, Elias Nehme, Moran Bercovici, Yoav Shechtman Science Advances, 2020 [paper] Leveraging PSF engineering it is possible to profile the topology of dynamic microsurface in 3D without scanning. |
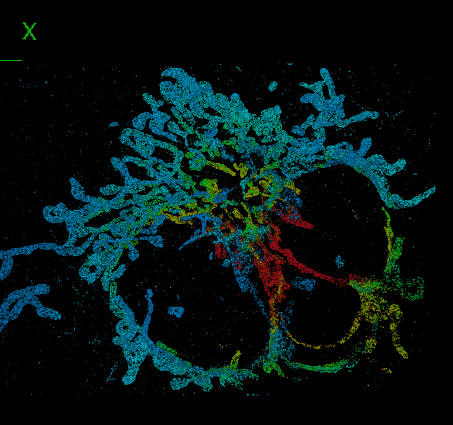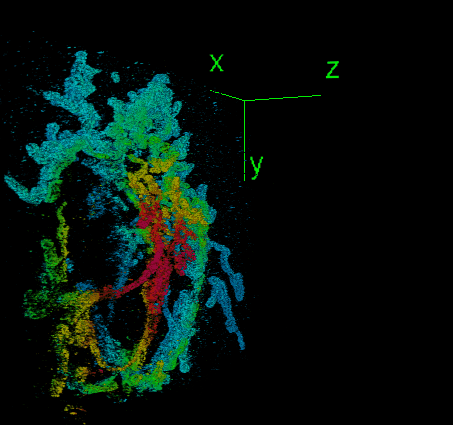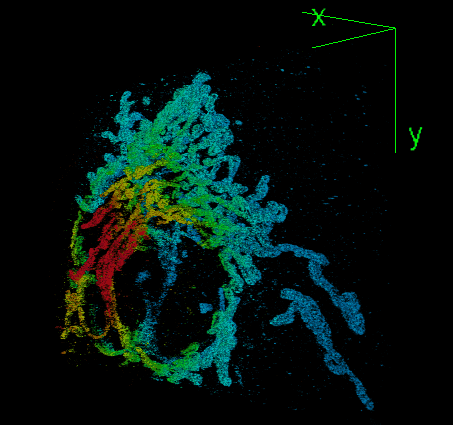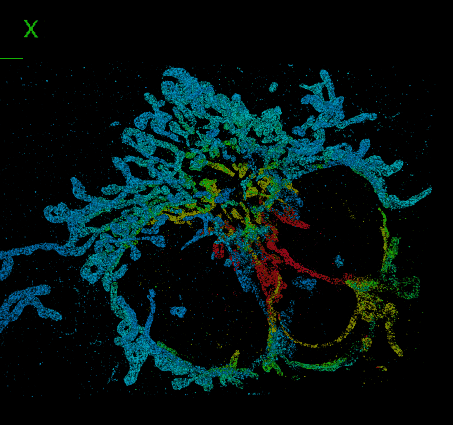 |
Elias Nehme, Daniel Freedman, Racheli Gordon, Boris Ferdman, Lucien E. Weiss, Onit Alalouf, Tal Naor, Reut Orange, Tomer Michaeli, Yoav Shechtman Nature Methods, 2020 (Best Poster Award at QBI 2019) [paper] / [code] / [video] We address the challenge of dense 3D localization from a single 2D image by harnessing neural networks for both acquisition design and image processing. |
 |
Boris Ferdman, Elias Nehme, Lucien E. Weiss, Reut Orange, Onit Alalouf, Yoav Shechtman Optics Express, 2020 [paper] / [code] Vectorial phase retreival facilitated by utilizing Wirtinger flow and vectorial optics. |
|
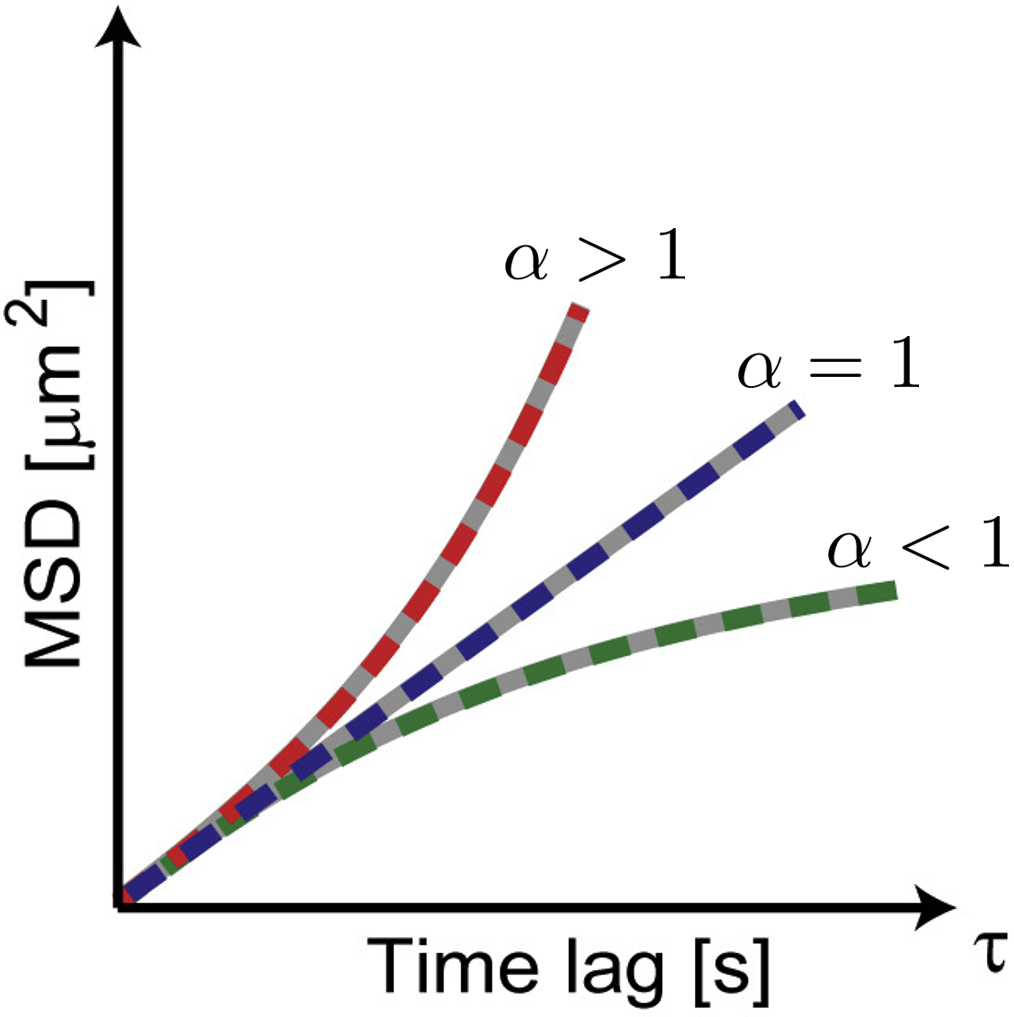
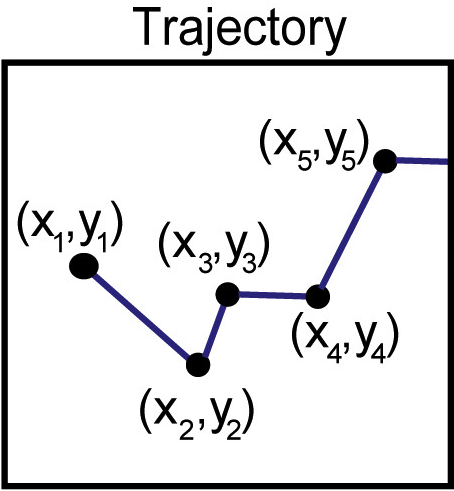
Naor Granik, Lucien E. Weiss, Elias Nehme, Maayan Levin, Michael Chein, Eran Perlson, Yael Roichman, Yoav Shechtman Biophysical Journal, 2019 [paper] / [code] Temporal convolutional networks can be employed to classify many short single-particle trajectories by diffusion type. |
 |
Elias Nehme, Lucien E. Weiss, Tomer Michaeli, Yoav Shechtman Optica, 2018 (Lev-Margulis Prize at ISM 2018, and Top cited article in Optica from 2018) [paper] / [code] Using a physical simulator we learn a neural network that achieves SotA performance in dense 2D localization microscopy. |
|
|
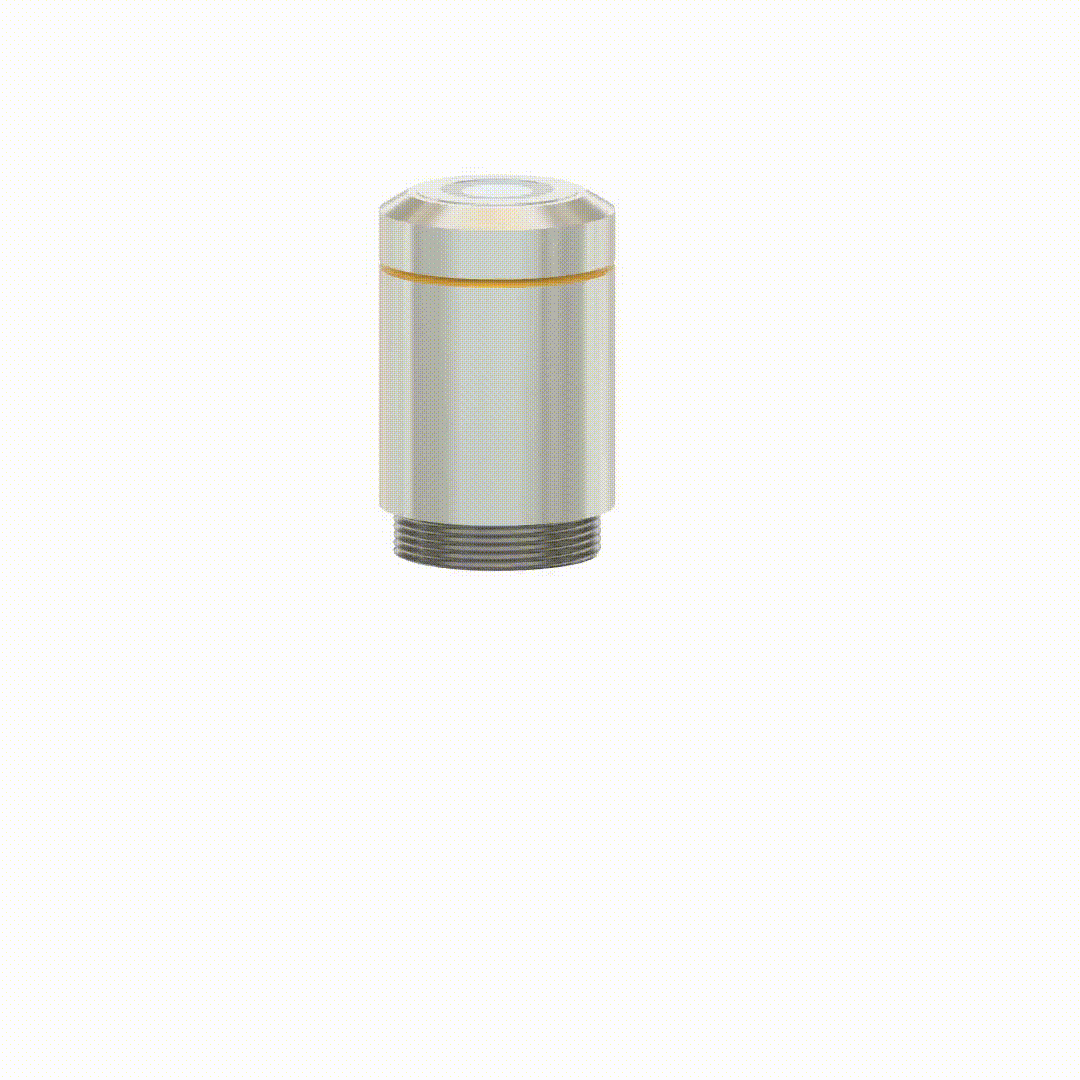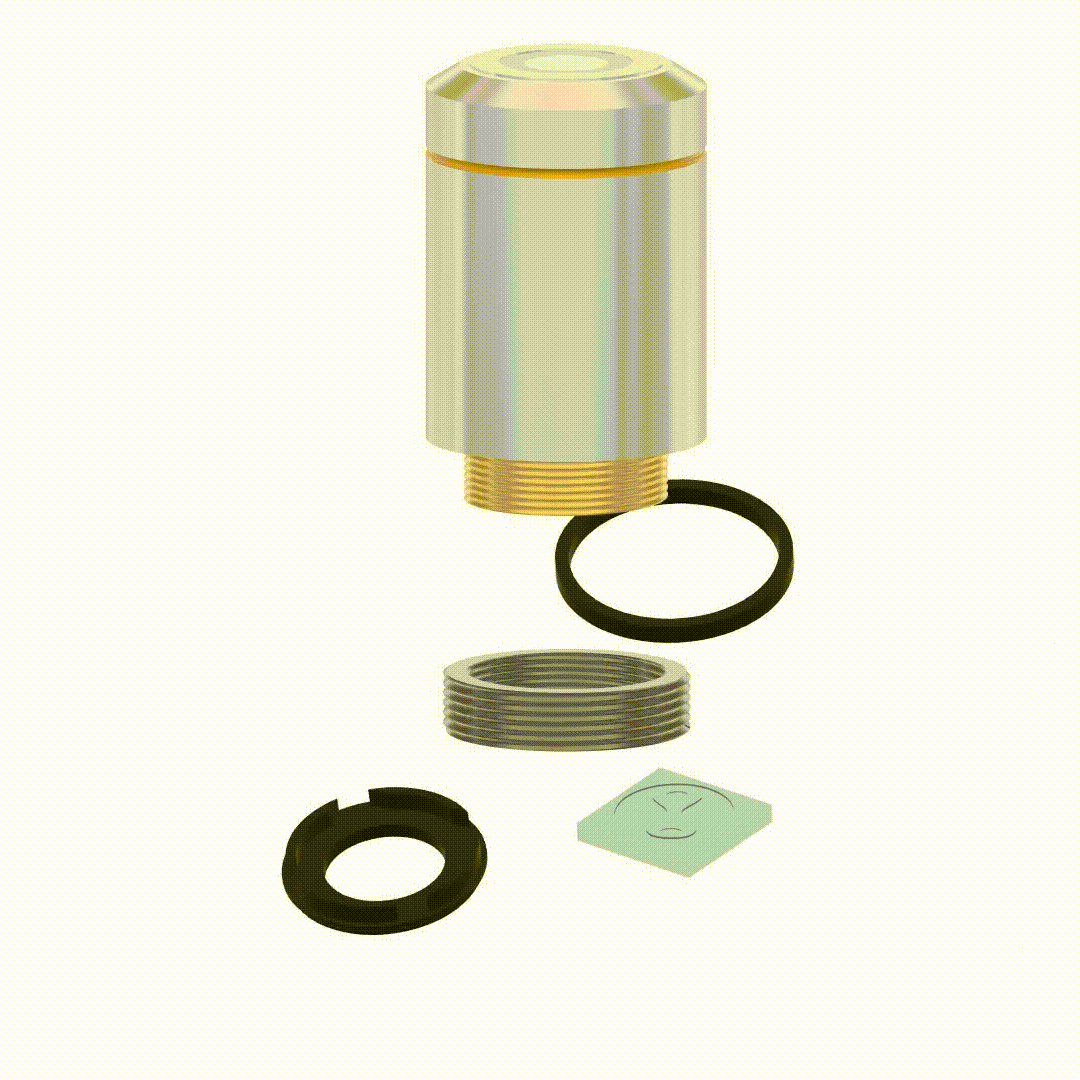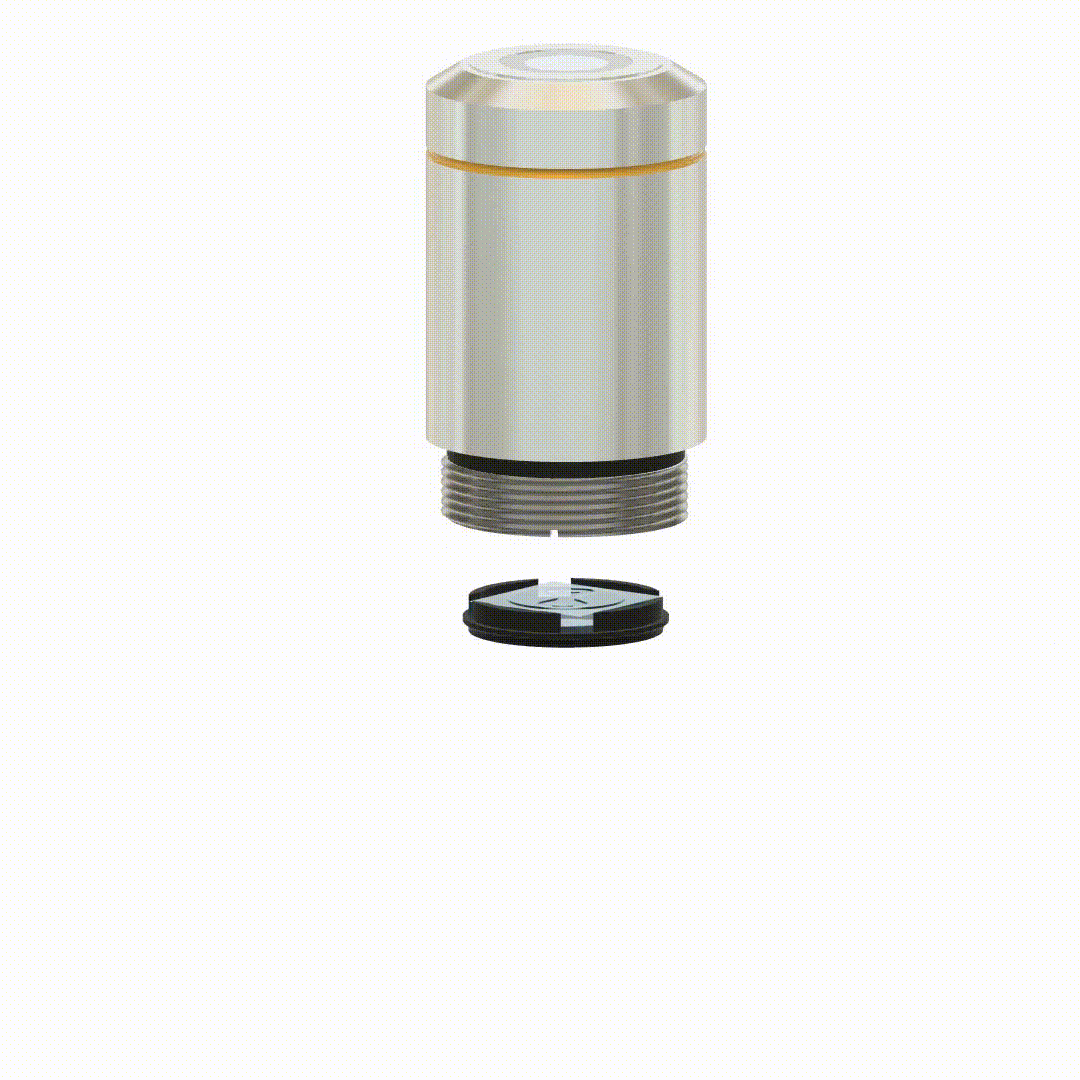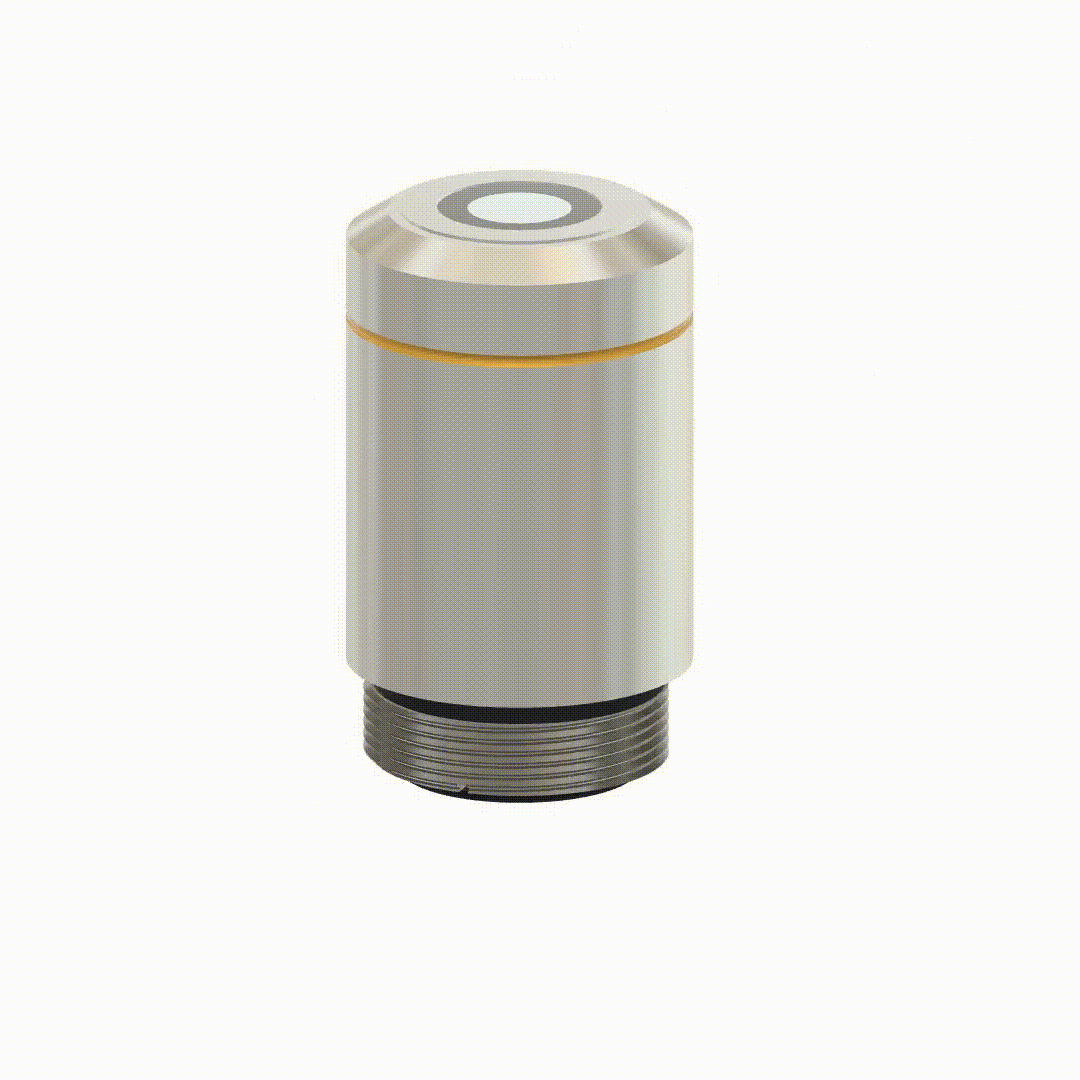 |
Yoav Shechtman, Boris Ferdman, Nadav Opatovski, Elias Nehme, Reut Orange-Kedem US Patent US20240272411A1, 2022 [patent] It is possible to modify objective lenses to perform compact wavefront modulation without a 4f system extension. |
|
|
 |
Teaching in English with Prof. Tomer Michaeli [EE-048954] Spring 2022 The course focuses on deep generative models for images describing practical strategies for learning high-dimensional probability distributions from samples. |
 |
Taught in English with Tal Daniel, Dahlia Urbach, Hila Manor, and Prof. Anat Levin [EE-046746] Spring 2021, Winter 2022 The course focuses on fundamental problems in computer vision describing practical solutions including state-of-the-art deep learning approaches. |
 |
Taught in English with Prof. Yoav Shechtman [BME-336547] Winter 2020 The course surveys recent computational imaging techniques co-designing optical sensors and data processing algorithms to advance imaging capabilities. |
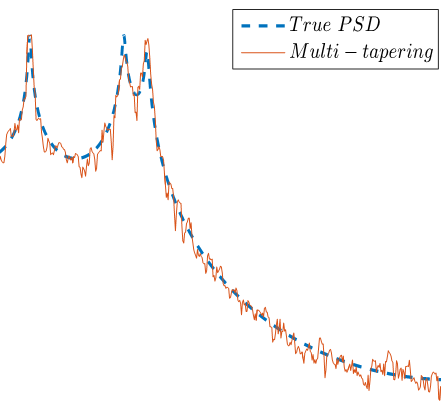 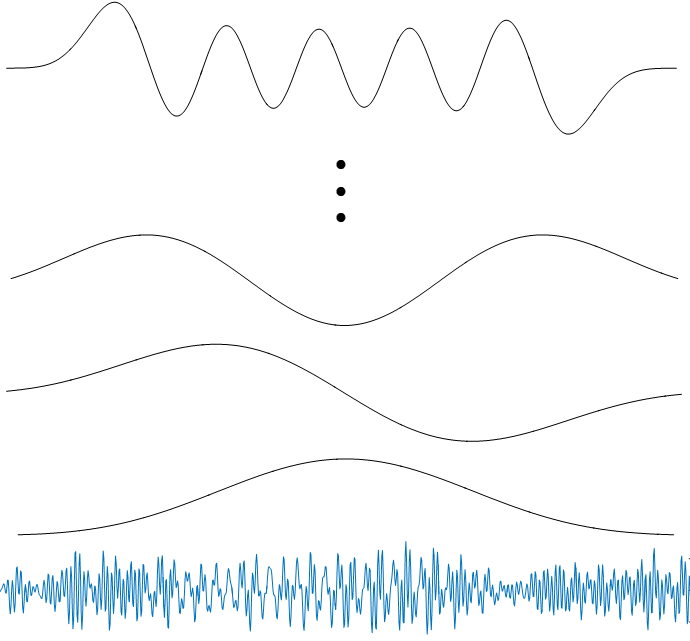
|
Taught in Hebrew with Prof. Yoav Shechtman [BME-336208] Spring 2018-2020 The course focuses on fundamental approaches in biological signal processing including linear filtering, correlation functions, non-stationary signals, and point processes. |
|
|
|
Taught in Hebrew with Prof. Yoav Shechtman Israel Ministry of Science and Technion-IIT Winter 2019-2020 Fundamentals of biosignal and bioimage processing delivered to electronics high school teachers. |
 
|
2018-present Reviewing in multiple scientific communities including journals (e.g. Optica, Nature Scientific Reports), and computer vision/machine learning conferences (e.g. CVPR, NeurIPS). |
|
Website credits to Jon Barron. |